
Complex wavelets for extended depth-of-field: a new method for the fusion of multichannel microscopy images. Deciphering dynamics of clathrin-mediated endocytosis in a living organism. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Flat clathrin lattices are dynamic actin-controlled hubs for clathrin-mediated endocytosis and signalling of specific receptors. Super-resolution video microscopy of live cells by structured illumination. Photostability of a fluorescent marker under pulsed excited-state depletion through stimulated emission. Stimulated emission depletion nanoscopy of living cells using SNAP-tag fusion proteins.

Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Light-induced cell damage in live-cell super-resolution microscopy. Wäldchen, S., Lehmann, J., Klein, T., van de Linde, S. Transfer learning in MIR: sharing learned latent representations for music audio classification and similarity. Stimulated emission depletion (STED)microscopy: from theory to practice. An iteration formula for Fredholm integral equations of the first kind. An iterative technique for the rectification of observed distributions. Bayesian-based iterative method of image restoration. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. PALM and STORM: into large fields and high-throughput microscopy with sCMOS detectors.
DIFFRACTION LIMIT CONFOCAL MICROSCOPY SOFTWARE
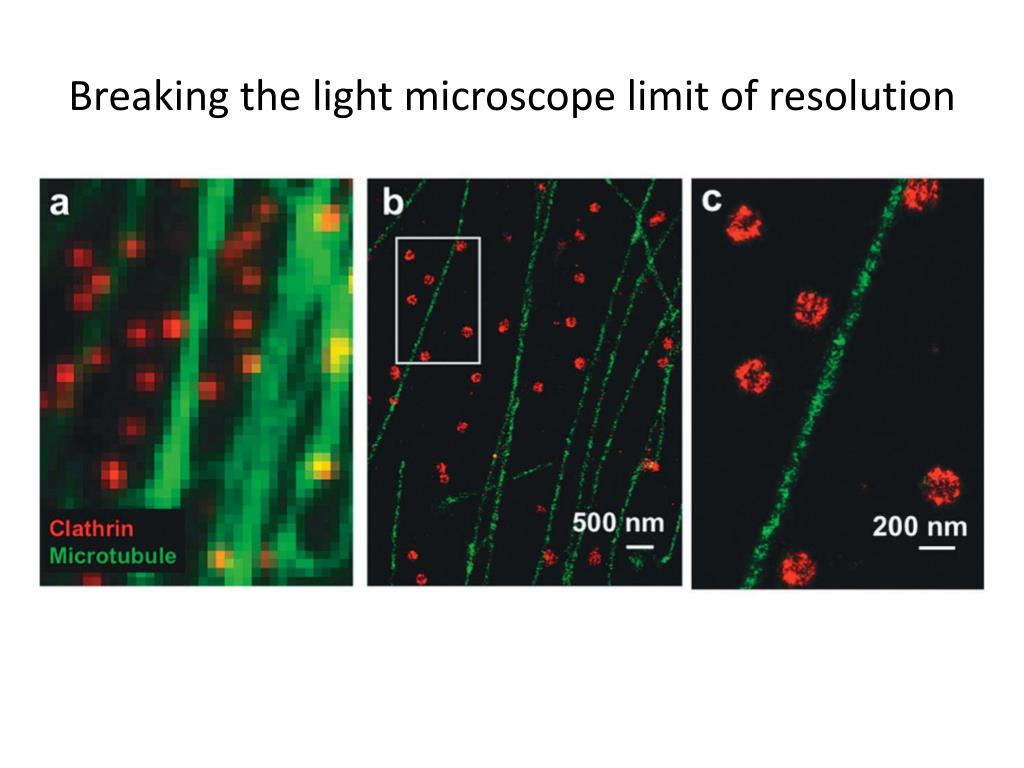
Quantitative evaluation of software packages for single-molecule localization microscopy. Quantitative mapping and minimization of super-resolution optical imaging artifacts. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Quantitative study of single molecule location estimation techniques. Fluorophore localization algorithms for super-resolution microscopy. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Imaging intracellular fluorescent proteins at nanometer resolution. The deep network rapidly outputs these super-resolved images, without any iterations or parameter search, and could serve to democratize super-resolution imaging.īetzig, E. We further demonstrate that total internal reflection fluorescence (TIRF) microscopy images of subcellular structures within cells and tissues can be transformed to match the results obtained with a TIRF-based structured illumination microscope. We also demonstrate cross-modality super-resolution, transforming confocal microscopy images to match the resolution acquired with a stimulated emission depletion (STED) microscope. Using this framework, we improve the resolution of wide-field images acquired with low-numerical-aperture objectives, matching the resolution that is acquired using high-numerical-aperture objectives. This data-driven approach does not require numerical modeling of the imaging process or the estimation of a point-spread-function, and is based on training a generative adversarial network (GAN) to transform diffraction-limited input images into super-resolved ones. We present deep-learning-enabled super-resolution across different fluorescence microscopy modalities.


 0 kommentar(er)
0 kommentar(er)
